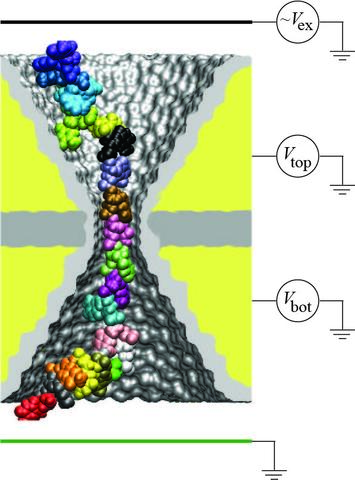
High-throughput technology for sequencing DNA has already provided invaluable information about the organization of the human genome and the common variations of the genome sequence among groups of individuals. To date, however, the high cost of whole genome-sequencing limits widespread use of this method in basic research and personal medicine. Using nanopores in synthetic silicon membranes for sequencing DNA can dramatic reduce the sequencing costs, as they enable, in principle, direct read out of the nucleotide sequence from the DNA strand via electric measurement. Although technology for manufacturing nanopores and using them for detecting single DNA molecules have already been demonstrated, a method for determining the DNA sequence with a single-nucleotide precision was lacking. In this study, we investigated the feasibility of sequencing DNA using an electric field in a nanopore that alternates periodically in time. Through molecular dynamics simulations, we demonstrated that back-and-forth motion of DNA in a 1-nm-diameter pore has a sequence-specific hysteresis that results in a detectable change of the electrostatic potential at the electrodes of the nanopore capacitor and in a sequence-specific drift of the DNA strand through the pore under an oscillating bias. On the basis of these observations, we proposed a method for detecting DNA sequences by modulating the duty cycle of the periodic alternating potential. When implemented technologically, this method can lead to major advances in healthcare, as it will make personal genomics an affordable reality. This study has been described in a report published in Nano Letters.