Refined Parameterization of Nonbonded Interactions Improves Conformational Sampling and Kinetics of Protein Folding Simulations
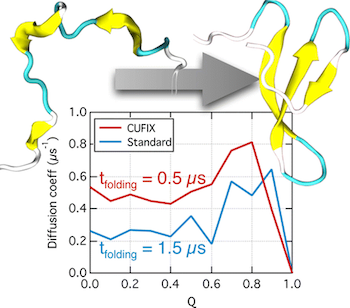
Recent advances in computational technology have enabled brute-force molecular dynamics (MD) simulations of protein folding using physics-based molecular force fields. The extensive sampling of protein conformations afforded by such simulations revealed, however, considerable compaction of the protein conformations in the unfolded state, which is inconsistent with experiment. Here, we show that a set of surgical corrections to nonbonded interactions between amine nitrogen–carboxylate oxygen and aliphatic carbon–carbon atom pairs can considerably improve the realism of protein folding simulations. Specifically, we show that employing our corrections in ∼500 μs all-atom replica-exchange MD simulations of the WW domain and villin head piece proteins increases the size of the denatured proteins’ conformations and does not destabilize the native conformations of the proteins. In addition to making the folded conformations a global minimum of the respective free energy landscapes at room temperature, our corrections also make the free energy landscape smoother, considerably accelerating the folding kinetics and, hence, reducing the computational expense of a protein folding simulation.