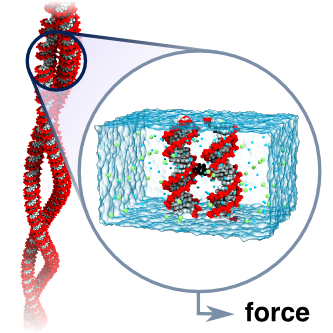
DNA is so famously known as the carrier of genetic information that the structural and dynamical aspects of the molecule are often neglected. However, most cellular processes that involve DNA cannot be understood without consideration of its interactions with other DNA and proteins. Such interactions can give rise self-assembled structures, for example DNA supercoils, which we strive to understand using a bottom-up approach by examining the most basic constituents of a larger more complex system. Thus, in collaboration with the groups of Ralf Seidel at the University of Technology in Dresden and Gero Wedemann at the University of Applied Sciences Stralsund, we have examined the interactions between DNA helices in plectonemic supercoils using magnetic tweezers, coarse-grained Monte Carlo and atomistic molecular dynamics simulations. Building on our previous work, our group characterized the effective forces between parallel DNA in monovalent electrolytes at different ion concentrations. Our simulations revealed that the force between the DNA molecules is much smaller than that predicted by the Debye-Hückel cylinder model and that continuum models fail to describe DNA electrostatics. We further demonstrate that DNA-DNA interactions in monovalent electrolyte are well described using this simple cylinder model provided a significant charge adaptation factor is employed. Application of this model in coarse-grained Monte Carlo simulations provided results in excellent agreement with experimentally obtained results over a wide range of concentrations. This work is described in a report that appeared in Physical Review Letters.