Modeling thermophoretic effects in solid-state nanopores
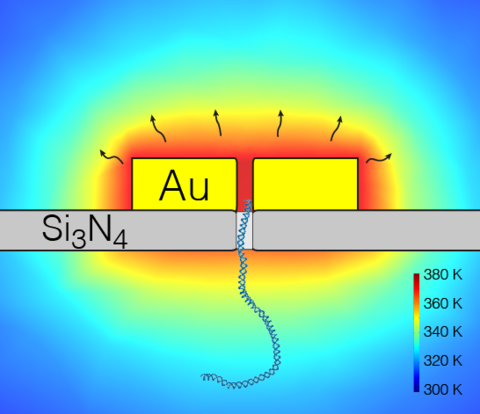
Local modulation of temperature has emerged as a new mechanism for regulation of molecular transport through nanopores. Predicting the effect of such modulations on nanopore transport requires simulation protocols capable of reproducing non-uniform temperature gradients observed in experiment. Conventional molecular dynamics (MD) method typically employs a single thermostat for maintaining a uniform distribution of temperature in the entire simulation domain, and, therefore, can not model local temperature variations. In this article, we describe a set of simulation protocols that enable modeling of nanopore systems featuring non-uniform distributions of temperature. First, we describe a method to impose a temperature gradient in all-atom MD simulations based on a boundary-driven non-equilibrium MD protocol. Then, we use this method to study the effect of temperature gradient on the distribution of ions in bulk solution (the thermophoretic effect). We show that DNA nucleotides exhibit differential response to the same temperature gradient. Next, we describe a method to directly compute the effective force of a thermal gradient on a prototypical biomolecule — a fragment of double-stranded DNA. Following that, we demonstrate an all-atom MD protocol for modeling thermophoretic effects in solid-state nanopores. We show that local heating of a nanopore volume can be used to regulate the nanopore ionic current. Finally, we show how continuum calculations can be coupled to a coarse-grained model of DNA to study the effect of local temperature modulation on electrophoretic motion of DNA through plasmonic nanopores. The computational methods described in this article are expected to find applications in rational design of temperature-responsive nanopore systems.
Electrophoretic flow of ions through a solid-state nanopore at 295 K
Electrophoretic flow of ions through a locally-heated solid-state nanopore heated up to 695 K.
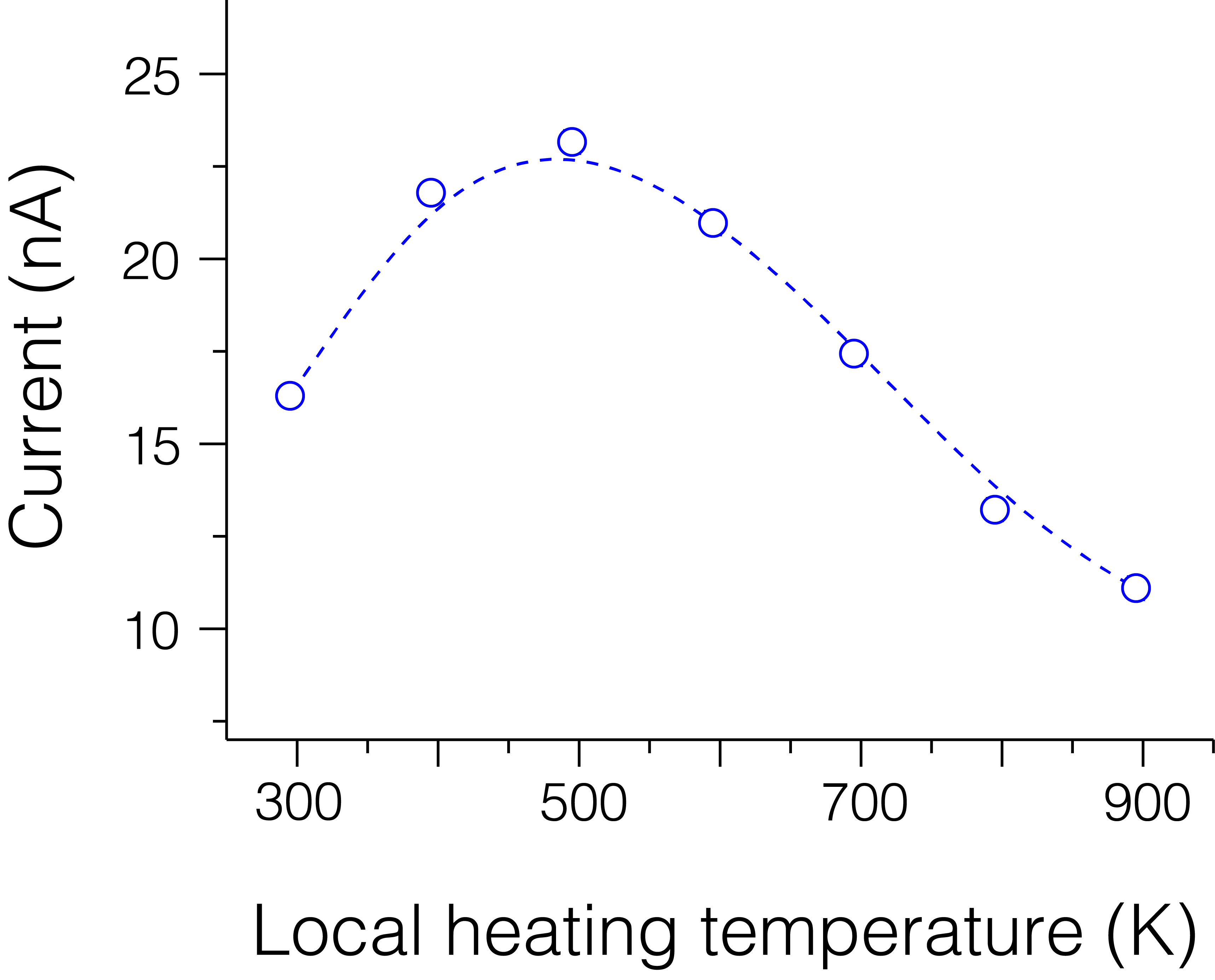
Ionic current through a solid-state Si3N4 nanopore as a function of the local-heating temperature under an applied bias of 1 V.
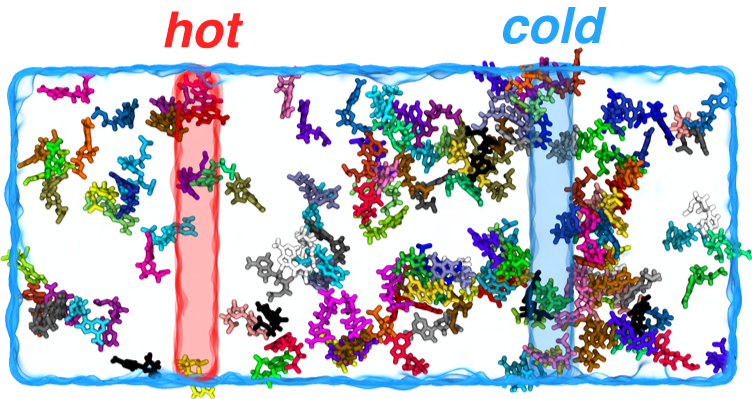
Positive thermophoresis of nucleotides: aggregation of nucleotides in the cooler region of a simulation box (marked as transparent blue).