Two Structural Scenarios for Protein Stabilization by PEG.
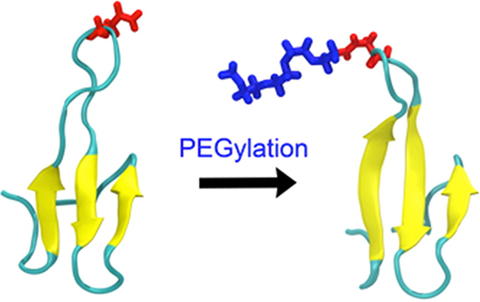
PEGylation, or addition of poly(ethylene glycol) chains to proteins, is widely used to improve delivery in pharmaceutical applications. Recent studies suggest that stabilization of a protein by PEG, and hence its proteolytic degradability, are sequence-dependent and require only short PEG chains. Here we connect stabilization by short PEG chains directly to the structural dynamics of the protein and PEG chain. We measured the stability of human Pin1 WW domain with PEG-4 at asparagine 19 for a full mutant cycle at two positions thought to influence PEG-protein interaction: Ser16Ala and Tyr23Phe. We then performed explicit solvent molecular dynamics simulations on all PEGylated and PEG-free mutants. The mutant cycle yields a non-additive stabilization effect where the pseudo wild type and double mutant are more stabilized relative to unPEGylated proteins than are the two single mutants. The simulation reveals why: the double mutant suffers loss of beta sheet structure, which PEGylation restores even though the PEG extends as a coil into the solvent. In contrast, in one of the single mutants PEG preferentially interacts with the protein surface while disrupting the interactions of its asparagine host with a nearby methionine side chain. Thus PEG attachment can stabilize a protein differentially depending on the local sequence, and either by interacting with the surface, or by extending into the solvent. A simulation with PEG-45 attached to asparagine 19 shows that PEG even can do both in the same context.